Building Infrared Proximity Sensors | NUS Electrical & Computer Engineering Principles & Practice Module EE1111B
Context:
This was a module that I signed up for as a non-university student in 2021. In it, we were taught the physics behind electrical circuits as well as electrical engineering with an Arduino. The final graded project for my batch was building a proximity sensor instead of a robotic car due to the COVID-19 pandemic.
Setup:

The key components are an infrared emitter and reciever pair. The reciever detects reflected infrared radiation and the Arduino measures the amount of voltage across both components.
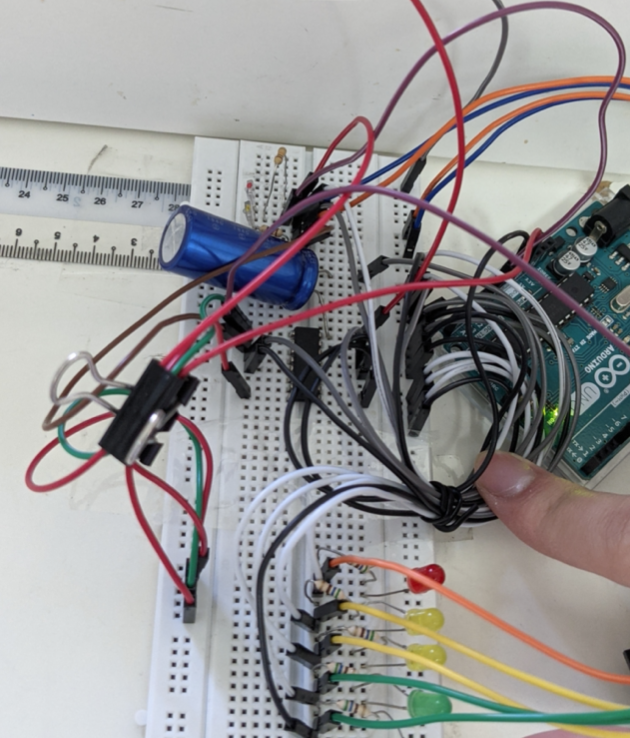
The emitter recieves current that is made consistent by the blue capcitator and is chopped up into small parts(pulse-width modulation) by the chip in the middle.
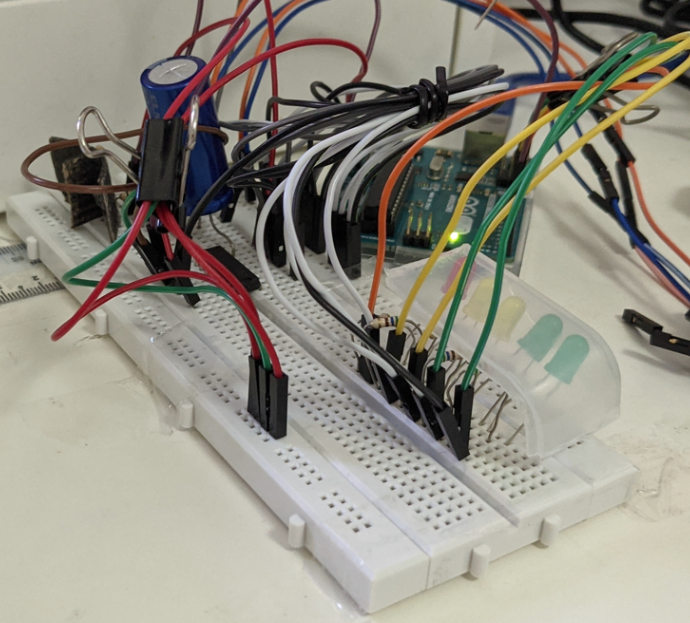
Here is the complete setup. The emitter and reciever pair are shielded with aluminium and black cardboard while a row of bulbs light up to indicate distance.
Code:
The setup is powered by 2 scripts. One is responsible for gathering voltage data and the other is used for testing functions that interpret the data.
A genetic algorithm written in Python was used to generate the function that calculates distance from data, where functions mutate, compete with one another, and reproduce if they are more accurate.
# An organism looks like this: A * B ^ C(x + D) + E, where
# A, B, C, D and E are the factors that make up an organism
# and x is a measured value like amplitudeVoltage
# stores the genepool
list_of_organisms = []
# a bunch of constants to be used in the genetic algorithm
number_of_organisms = 1000 # size of genepool
number_of_factors = 5 # how many factors define an organism
step = 0.5 # mutation rate
#code for randomly generating the first organisms
for i in range(number_of_organisms): # generate number_of_organisms number of organisms
organism = [] # a single blank organisms
for i in range(number_of_factors): # generates 5 random factors for the organism
organism.append(random.uniform(0, 10))
organism.append(0) # adds a sixth value, which is the organism's fitness value
list_of_organisms.append(organism) # add the newly created organism to the genepool
curr_best_organism = list_of_organisms[0] # the fittest organism so far,
for i in range(1, 10000000):
# Evaluate fitness
for organism in list_of_organisms:
fitness_value = 0
#go through every data point from the data to evaluate fitness of organism
for distance_value, measurements in data.items():
# the fitness value is the sum of the diffference between the actual distance_value and the distance calculated from the equation A * B ^ C(x + D) + E
fitness_value += (distance_value - (organism[0] * organism[1] ** (organism[2] * (measurements[2] + organism[3])) + organism[4]))
#fitness_value += abs(measurements[2] - organism[0] * math.log(abs(distance_value), abs(organism[1])))
organism[number_of_factors] = abs(fitness_value/len(target_data))
###############
# sort the organisms by their fitness
list_of_organisms = sorted(list_of_organisms,key=lambda x: x[number_of_factors])
# if the fittest organism is fitter than the organism stored in curr_best_organism,
if list_of_organisms[0][number_of_factors] < curr_best_organism[number_of_factors]:
curr_best_organism = list_of_organisms[0] # store that fittest organism in curr_best_organism
# print the generation number, curr_best_organism's fitness and the factors that define it
print("Generation " + str(i), curr_best_organism[number_of_factors], curr_best_organism[0:number_of_factors])
###############
# Sort organisms by fitness, replicate and mutate them
# sorted_list = sorted(list_not_sorted, key=lambda x:x[2])
next_list_of_organisms = [] # create a new blank genepool
for i1 in range(number_of_organisms//2): # go through the fittest half of the previous genepool
copy_of_organism = list_of_organisms[i1] # copy over each individual organism to be mainpulated
next_list_of_organisms.append(copy_of_organism) # copy over that organism into the new genepool
next_list_of_organisms.append([copy_of_organism[0] + random.uniform(-step, step), # mutate each of the 5 factors, and then add the mutated organism into the genepool
copy_of_organism[1] + random.uniform(-step, step),
copy_of_organism[2] + random.uniform(-step, step),
copy_of_organism[3] + random.uniform(-step, step),
copy_of_organism[4] + random.uniform(-step, step),
copy_of_organism[number_of_factors]])
list_of_organisms = next_list_of_organisms # save the new genepool into list_of_organisms